Abstract
Introduction: Although outcomes for pediatric acute lymphoblastic leukemia (ALL) have improved dramatically, five-year survival remains dismal for those cases which relapse. Initial response to therapy, measured by minimal residual disease (MRD) at end-induction, is the most important predictor of outcome. However, approximately half of relapses occur in MRD-negative patients. We hypothesized that global metabolomic profiling of diagnostic bone marrow plasma could improve prediction of MRD positivity and relapse when combined with clinical features, while also providing insights into the biologic features of these chemotherapy-resistant groups.
Methods: The study population (n=100) included children age 0-18 years with newly diagnosed ALL treated at Texas Children's Cancer Center. The cohort included 39 cases with MRD ≥ 0.01% at end-induction. Ninety-four children were followed for at least 3 years from diagnosis, of whom 23 relapsed. Diagnostic bone marrow plasma specimens were obtained with informed consent and subjected to metabolomic profiling via UPLC-MS/MS by Metabolon. Following compound identification and correction for batch effects, each compound was scaled such that the median concentration was one and missing values were imputed with the minimum detected concentration.
Analysis was limited to 712 named compounds that were detected in at least two samples. We used Welch's two-sample t-test to compute p-values comparing mean concentrations in MRD positive vs. negative and relapsed vs. non-relapsed plasma and chi-squared test to assess differences in the frequency of detection. We then generated volcano plots to visualize the significance of each metabolite.
Compounds demonstrating significance at the p < 0.001 level were then evaluated in logistic regression models for MRD and relapse, and the best predictors were obtained by backward elimination. Finally, we assessed the performance of models to predict MRD and relapse using (1) clinical features alone (immunophenotype, cytogenetics and National Cancer Institute risk category, plus end-induction MRD when predicting relapse); (2) metabolomic data alone; and (3) clinical features + metabolomic data. DeLong's test for correlated receiver operating characteristics curves was used to compare model performance.
Results: In the assessment of individual metabolites, a higher concentration of pyruvate was associated with an increased odds ratio (OR) of MRD positivity (p = 0.005). Conversely, a significantly reduced OR for MRD was identified for arachidonylglycerol (p = 0.01). Reduced ORs for relapse were identified for increasing concentrations of two compounds involved in amino acid metabolism: picolinic acid and N-carboxyethylphenylalanine (p = 0.04 and 0.02, respectively), as well as for the vitamin E derivative gamma-CEHC (p = 0.03). Several glycerophospholipids demonstrated inverse associations with MRD positivity and relapse.
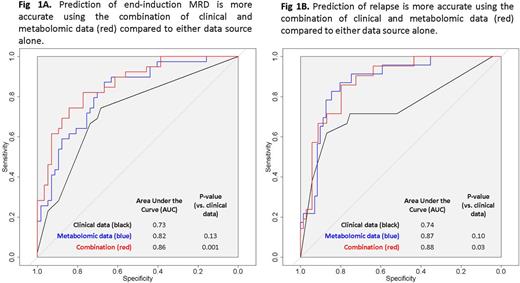
Metabolomic profiling significantly improved prediction of both MRD and relapse as compared to clinical features alone (p = 0.001 and p = 0.03 respectively; Figure 1). Incorporating metabolomic data gave an absolute improvement in sensitivity of 8% for MRD and 19% for relapse.
Conclusions: We demonstrate that global metabolomic profiling improves sensitivity and specificity when predicting risk of MRD positivity and relapse in ALL. Differences in pyruvate and lipid concentrations suggest metabolic changes associated with chemoresistance or disease recurrence which may be therapeutically targetable. If validated, these metabolomic biomarkers may improve clinical risk prediction and further our understanding of disease biology.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal